Preprocessing for numerical features#
In this notebook, we still use numerical features only.
Here we introduce these new aspects:
an example of preprocessing, namely scaling numerical variables;
using a scikit-learn pipeline to chain preprocessing and model training.
Data preparation#
First, let’s load the full adult census dataset.
import pandas as pd
adult_census = pd.read_csv("../datasets/adult-census.csv")
We now drop the target from the data we use to train our predictive model.
target_name = "class"
target = adult_census[target_name]
data = adult_census.drop(columns=target_name)
Then, we select only the numerical columns, as seen in the previous notebook.
numerical_columns = ["age", "capital-gain", "capital-loss", "hours-per-week"]
data_numeric = data[numerical_columns]
Finally, we can divide our dataset into a train and test sets.
from sklearn.model_selection import train_test_split
data_train, data_test, target_train, target_test = train_test_split(
data_numeric, target, random_state=42
)
Model fitting with preprocessing#
A range of preprocessing algorithms in scikit-learn allow us to transform the input data before training a model. In our case, we will standardize the data and then train a new logistic regression model on that new version of the dataset.
Let’s start by printing some statistics about the training data.
data_train.describe()
| age | capital-gain | capital-loss | hours-per-week | |
|---|---|---|---|---|
| count | 36631.000000 | 36631.000000 | 36631.000000 | 36631.000000 |
| mean | 38.642352 | 1087.077721 | 89.665311 | 40.431247 |
| std | 13.725748 | 7522.692939 | 407.110175 | 12.423952 |
| min | 17.000000 | 0.000000 | 0.000000 | 1.000000 |
| 25% | 28.000000 | 0.000000 | 0.000000 | 40.000000 |
| 50% | 37.000000 | 0.000000 | 0.000000 | 40.000000 |
| 75% | 48.000000 | 0.000000 | 0.000000 | 45.000000 |
| max | 90.000000 | 99999.000000 | 4356.000000 | 99.000000 |
We see that the dataset’s features span across different ranges. Some algorithms make some assumptions regarding the feature distributions and normalizing features is usually helpful to address such assumptions.
Tip
Here are some reasons for scaling features:
Models that rely on the distance between a pair of samples, for instance k-nearest neighbors, should be trained on normalized features to make each feature contribute approximately equally to the distance computations.
Many models such as logistic regression use a numerical solver (based on gradient descent) to find their optimal parameters. This solver converges faster when the features are scaled, as it requires less steps (called iterations) to reach the optimal solution.
Whether or not a machine learning model requires scaling the features depends on the model family. Linear models such as logistic regression generally benefit from scaling the features while other models such as decision trees do not need such preprocessing (but would not suffer from it).
We show how to apply such normalization using a scikit-learn transformer
called StandardScaler. This transformer shifts and scales each feature
individually so that they all have a 0-mean and a unit standard deviation.
We recall that transformers are estimators that have a transform method.
We now investigate different steps used in scikit-learn to achieve such a transformation of the data.
First, one needs to call the method fit in order to learn the scaling from
the data.
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(data_train)
StandardScaler()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
StandardScaler()
The fit method for transformers is similar to the fit method for
predictors. The main difference is that the former has a single argument (the
data matrix), whereas the latter has two arguments (the data matrix and the
target).
In this case, the algorithm needs to compute the mean and standard deviation for each feature and store them into some NumPy arrays. Here, these statistics are the model states.
Note
The fact that the model states of this scaler are arrays of means and standard
deviations is specific to the StandardScaler. Other scikit-learn
transformers may compute different statistics and store them as model states,
in a similar fashion.
We can inspect the computed means and standard deviations.
scaler.mean_
array([ 38.64235211, 1087.07772106, 89.6653108 , 40.43124676])
scaler.scale_
array([ 13.72556083, 7522.59025606, 407.10461772, 12.42378265])
Note
scikit-learn convention: if an attribute is learned from the data, its name
ends with an underscore (i.e. _), as in mean_ and scale_ for the
StandardScaler.
Scaling the data is applied to each feature individually (i.e. each column in the data matrix). For each feature, we subtract its mean and divide by its standard deviation.
Once we have called the fit method, we can perform data transformation by
calling the method transform.
data_train_scaled = scaler.transform(data_train)
data_train_scaled
array([[ 0.17177061, -0.14450843, 5.71188483, -2.28845333],
[ 0.02605707, -0.14450843, -0.22025127, -0.27618374],
[-0.33822677, -0.14450843, -0.22025127, 0.77019645],
...,
[-0.77536738, -0.14450843, -0.22025127, -0.03471139],
[ 0.53605445, -0.14450843, -0.22025127, -0.03471139],
[ 1.48319243, -0.14450843, -0.22025127, -2.69090725]])
Let’s illustrate the internal mechanism of the transform method and put it
to perspective with what we already saw with predictors.
The transform method for transformers is similar to the predict method for
predictors. It uses a predefined function, called a transformation
function, and uses the model states and the input data. However, instead of
outputting predictions, the job of the transform method is to output a
transformed version of the input data.
Finally, the method fit_transform is a shorthand method to call successively
fit and then transform.
In scikit-learn jargon, a transformer is defined as an estimator (an
object with a fit method) supporting transform or fit_transform.
data_train_scaled = scaler.fit_transform(data_train)
data_train_scaled
array([[ 0.17177061, -0.14450843, 5.71188483, -2.28845333],
[ 0.02605707, -0.14450843, -0.22025127, -0.27618374],
[-0.33822677, -0.14450843, -0.22025127, 0.77019645],
...,
[-0.77536738, -0.14450843, -0.22025127, -0.03471139],
[ 0.53605445, -0.14450843, -0.22025127, -0.03471139],
[ 1.48319243, -0.14450843, -0.22025127, -2.69090725]])
By default, all scikit-learn transformers output NumPy arrays. Since
scikit-learn 1.2, it is possible to set the output to be a pandas dataframe,
which makes data exploration easier as it preserves the column names. The
method set_output controls this behaviour. Please refer to this example
from the scikit-learn
documentation
for more options to configure the output of transformers.
scaler = StandardScaler().set_output(transform="pandas")
data_train_scaled = scaler.fit_transform(data_train)
data_train_scaled.describe()
| age | capital-gain | capital-loss | hours-per-week | |
|---|---|---|---|---|
| count | 3.663100e+04 | 3.663100e+04 | 3.663100e+04 | 3.663100e+04 |
| mean | -2.273364e-16 | 3.530310e-17 | 3.840667e-17 | 1.844684e-16 |
| std | 1.000014e+00 | 1.000014e+00 | 1.000014e+00 | 1.000014e+00 |
| min | -1.576792e+00 | -1.445084e-01 | -2.202513e-01 | -3.173852e+00 |
| 25% | -7.753674e-01 | -1.445084e-01 | -2.202513e-01 | -3.471139e-02 |
| 50% | -1.196565e-01 | -1.445084e-01 | -2.202513e-01 | -3.471139e-02 |
| 75% | 6.817680e-01 | -1.445084e-01 | -2.202513e-01 | 3.677425e-01 |
| max | 3.741752e+00 | 1.314865e+01 | 1.047970e+01 | 4.714245e+00 |
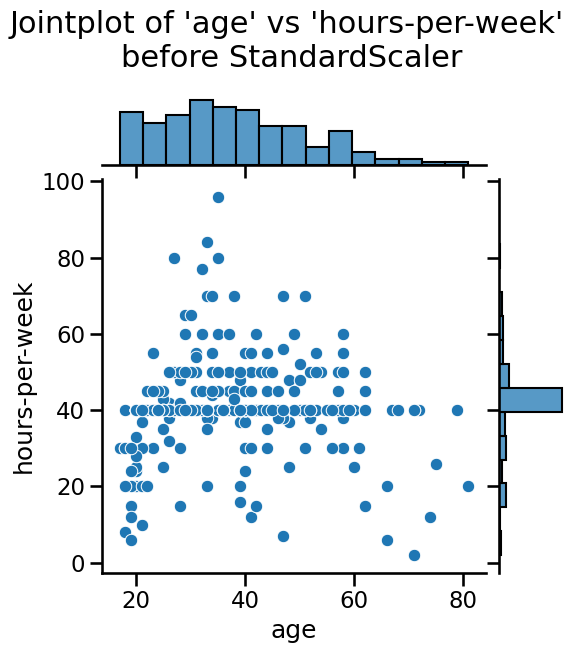
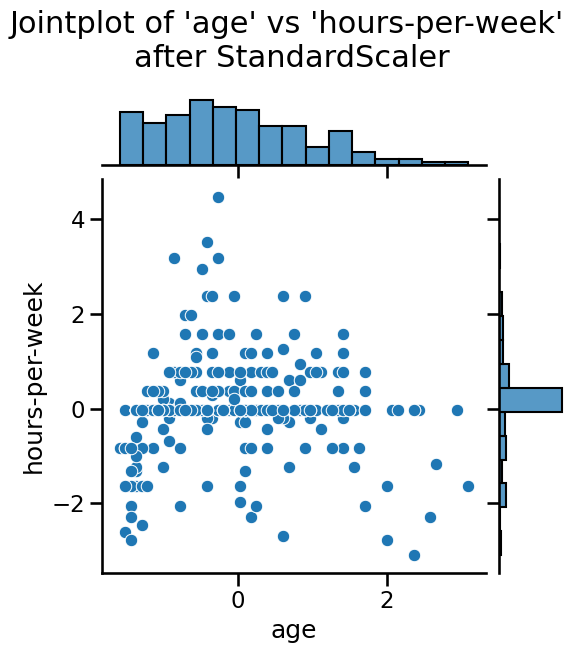
Notice that the mean of all the columns is close to 0 and the standard
deviation in all cases is close to 1. We can also visualize the effect of
StandardScaler using a jointplot to show both the histograms of the
distributions and a scatterplot of any pair of numerical features at the same
time. We can observe that StandardScaler does not change the structure of
the data itself but the axes get shifted and scaled.
Note to instructor: We strongly advise to copy-paste below code and discuss the visualization with your audience. We discourage using live-coding here, because you will spend a lot of time fiddling with the jointplot, time which we can better use to introduce machine learning.
import matplotlib.pyplot as plt
import seaborn as sns
# number of points to visualize to have a clearer plot
num_points_to_plot = 300
sns.jointplot(
data=data_train[:num_points_to_plot],
x="age",
y="hours-per-week",
marginal_kws=dict(bins=15),
)
plt.suptitle(
"Jointplot of 'age' vs 'hours-per-week' \nbefore StandardScaler", y=1.1
)
sns.jointplot(
data=data_train_scaled[:num_points_to_plot],
x="age",
y="hours-per-week",
marginal_kws=dict(bins=15),
)
_ = plt.suptitle(
"Jointplot of 'age' vs 'hours-per-week' \nafter StandardScaler", y=1.1
)
We can easily combine sequential operations with a scikit-learn Pipeline,
which chains together operations and is used as any other classifier or
regressor. The helper function make_pipeline creates a Pipeline: it
takes as arguments the successive transformations to perform, followed by the
classifier or regressor model.
import time
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import make_pipeline
model = make_pipeline(StandardScaler(), LogisticRegression())
model
Pipeline(steps=[('standardscaler', StandardScaler()),
('logisticregression', LogisticRegression())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('standardscaler', StandardScaler()),
('logisticregression', LogisticRegression())])StandardScaler()
LogisticRegression()
The make_pipeline function did not require us to give a name to each step.
Indeed, it was automatically assigned based on the name of the classes
provided; a StandardScaler step is named "standardscaler" in the resulting
pipeline. We can check the name of each steps of our model:
model.named_steps
{'standardscaler': StandardScaler(),
'logisticregression': LogisticRegression()}
This predictive pipeline exposes the same methods as the final predictor:
fit and predict (and additionally predict_proba, decision_function, or
score).
model.fit(data_train, target_train)
Pipeline(steps=[('standardscaler', StandardScaler()),
('logisticregression', LogisticRegression())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('standardscaler', StandardScaler()),
('logisticregression', LogisticRegression())])StandardScaler()
LogisticRegression()
We can represent the internal mechanism of a pipeline when calling fit by
the following diagram:
When calling model.fit, the method fit_transform from each underlying
transformer (here a single transformer) in the pipeline is called to:
learn their internal model states
transform the training data. Finally, the preprocessed data are provided to train the predictor.
To predict the targets given a test set, one uses the predict method.
predicted_target = model.predict(data_test)
predicted_target[:5]
array([' <=50K', ' <=50K', ' >50K', ' <=50K', ' <=50K'], dtype=object)
Let’s show the underlying mechanism:
The method transform of each transformer (here a single transformer) is
called to preprocess the data. Note that there is no need to call the fit
method for these transformers because we are using the internal model states
computed when calling model.fit. The preprocessed data is then provided to
the predictor that outputs the predicted target by calling its method
predict.
As a shorthand, we can check the score of the full predictive pipeline calling
the method model.score. Thus, let’s check the
generalization performance of such a predictive pipeline.
model_name = model.__class__.__name__
score = model.score(data_test, target_test)
print(f"The accuracy using a {model_name} is {score:.3f} ")
The accuracy using a Pipeline is 0.807
We can compare the pipeline using scaling and logistic regression with using a logistic regression model without scaling like we did before. We will not go into how to do this comparison with Python in sake of time, instead we directly give you the results:
The accuracy using a Pipeline is 0.807 with a fitting time of 0.043 seconds in 9 iterations The accuracy using a LogisticRegression is 0.807 with a fitting time of 0.110 seconds in 60 iterations
We see that scaling the data before training the logistic regression was beneficial in terms of computational performance. Indeed, the number of iterations decreased as well as the training time. The generalization performance did not change since both models converged.
Warning
Working with non-scaled data will potentially force the algorithm to iterate
more as we showed in the example above. There is also the catastrophic
scenario where the number of required iterations is larger than the maximum
number of iterations allowed by the predictor (controlled by the max_iter)
parameter. Therefore, before increasing max_iter, make sure that the data
are well scaled.
In this notebook we:
saw the importance of scaling numerical variables;
used a pipeline to chain scaling and logistic regression training.

